A New Tool for Mutational Signature Analysis
Hu and Doga describe MuSiCal (Mutational Signature Calculator) for analysis of somatic mutations (Nat Genetics 2024)
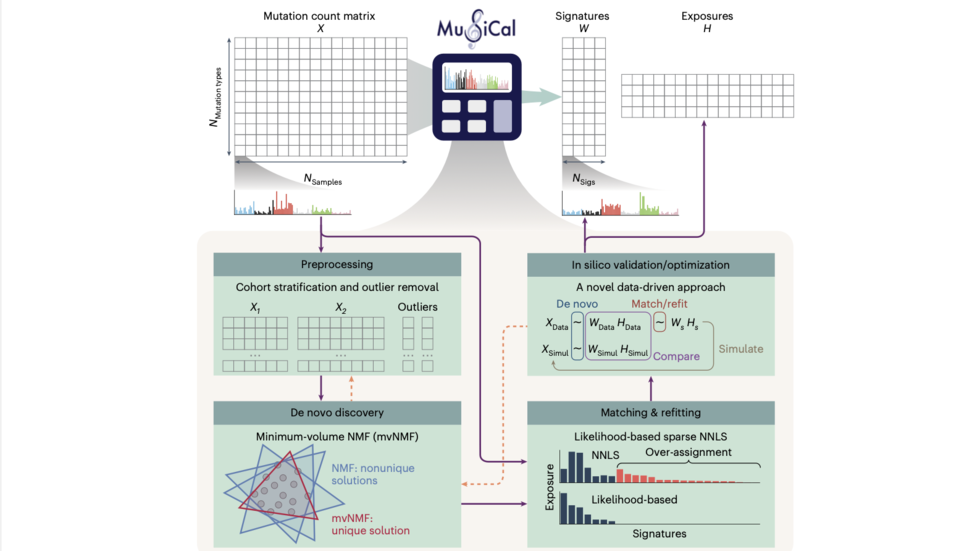
Hu and Doga describe MuSiCal (Mutational Signature Calculator) for analysis of somatic mutations (Nat Genetics 2024)
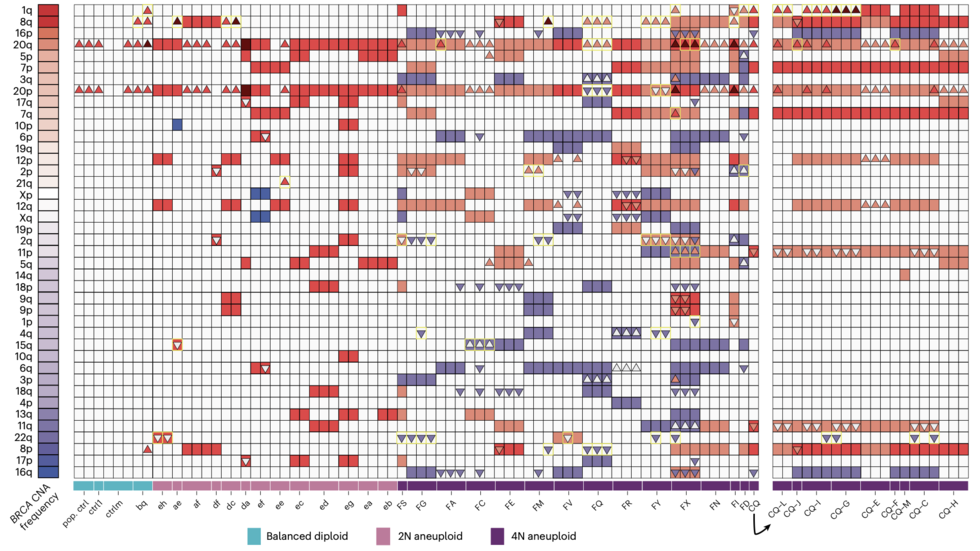
Chromosome evolution screens using single cell genome sequencing (Emma/Jake, Nat Genetics, 2024)
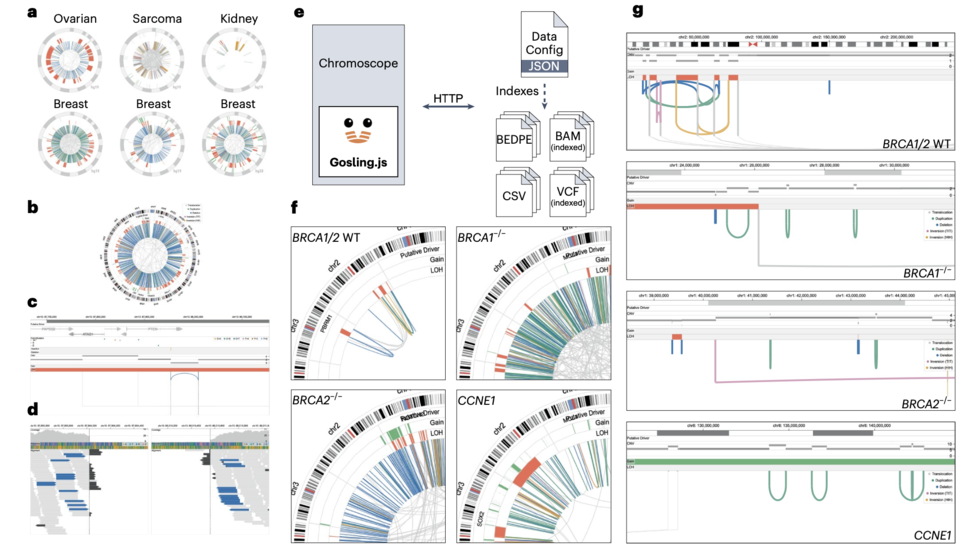
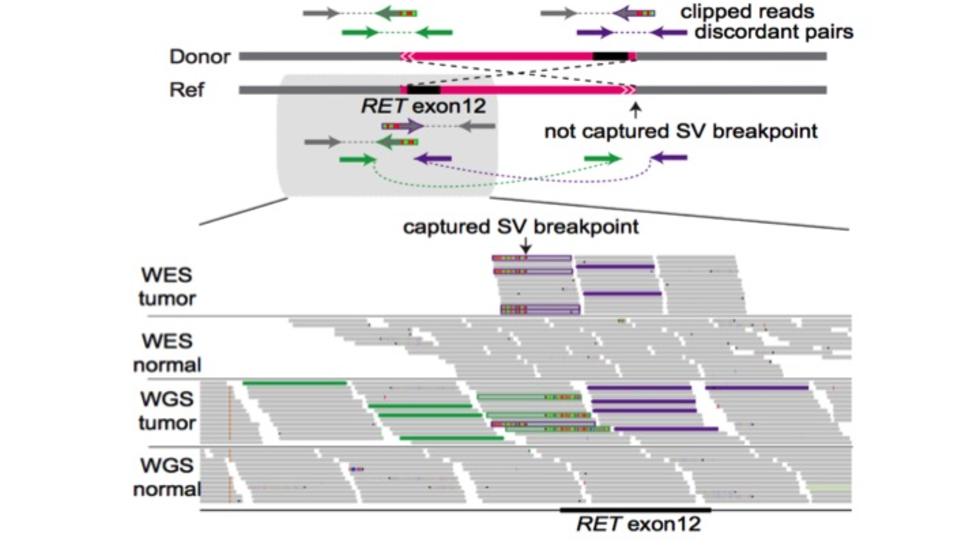
Interactive multiscale visualization for structural variation by Sehi and Dominik (Nat Methods, 2023)
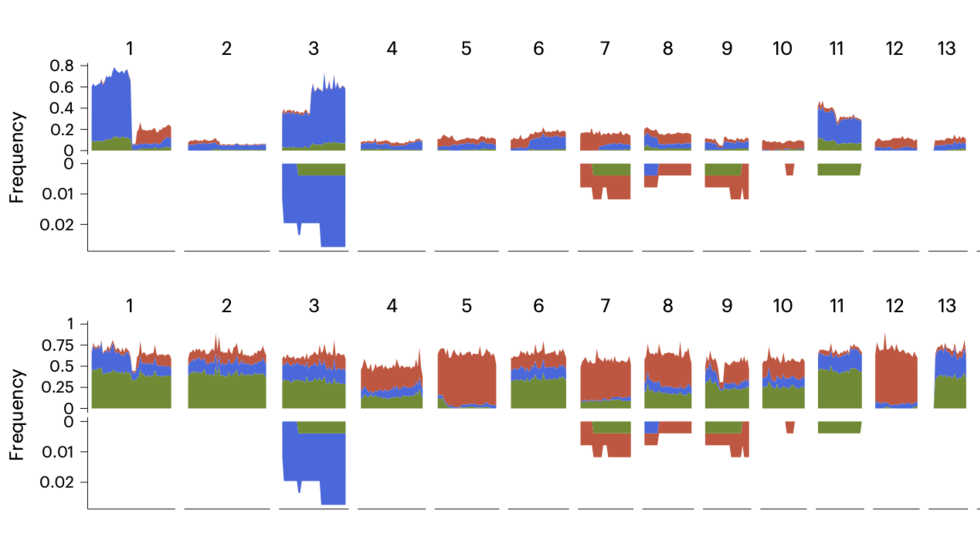
A pan-tissue survey of mosaic chromosomal alterations in 948 individuals (Teng/Kharchenko Lab, Nat Genetics, 2023)
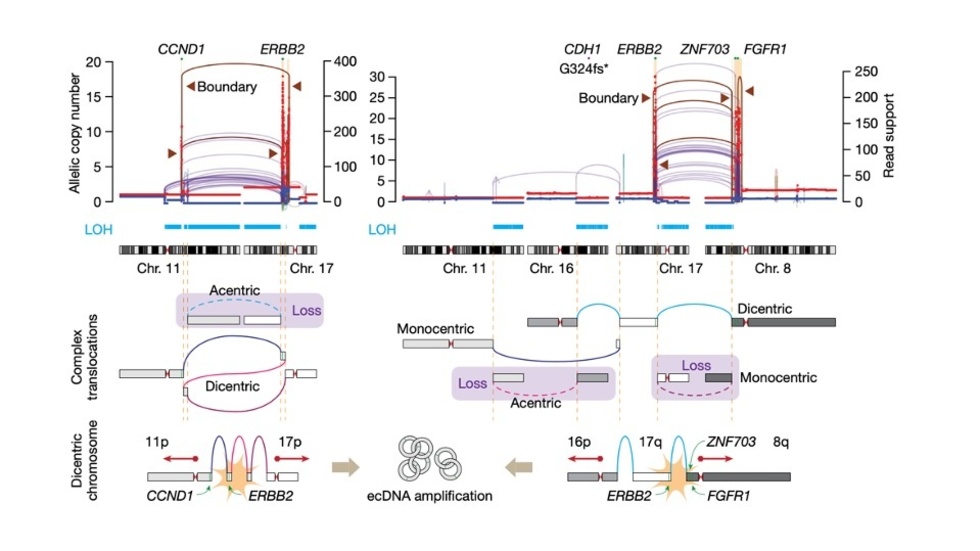
Jake discovers Translocation-Bridge Amplification that generates focal amplifications (Nature, 2023)
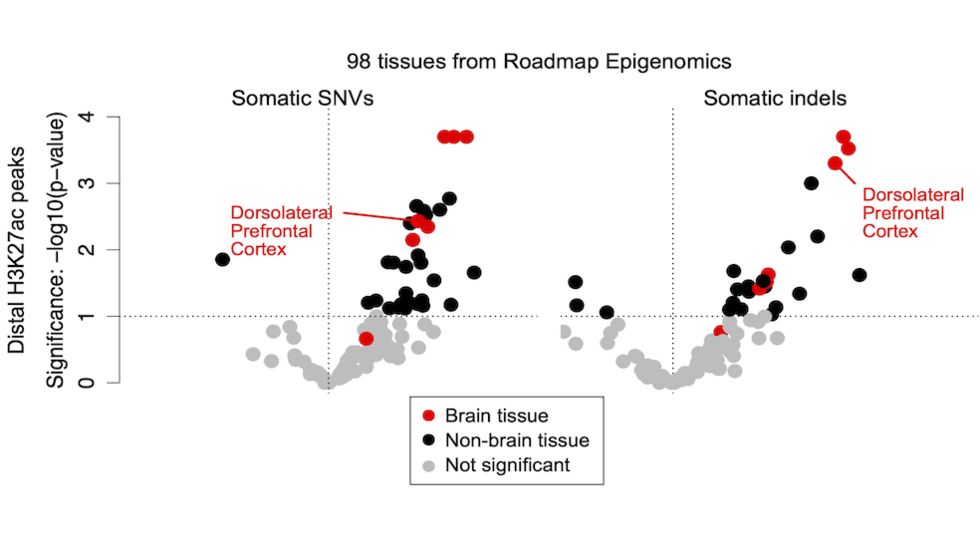
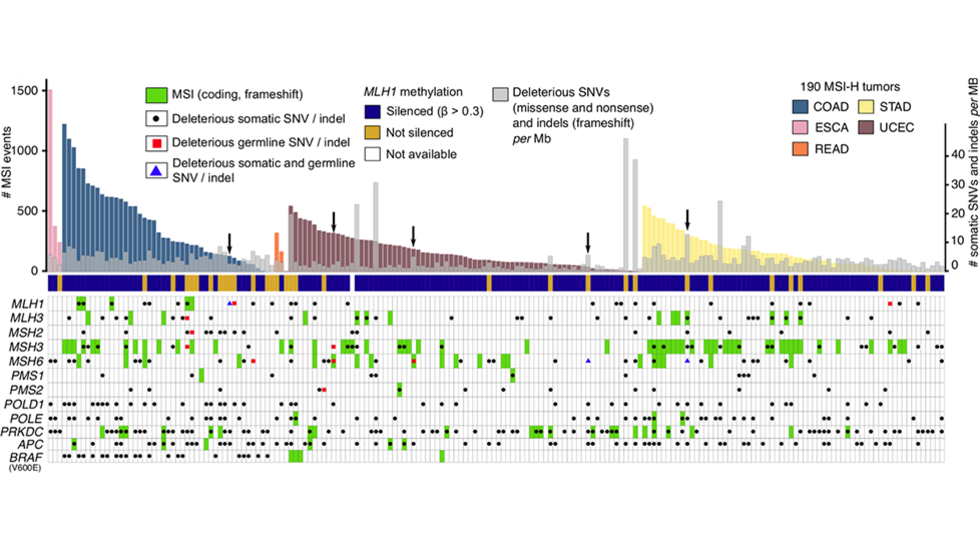
Joe finds enrichment of somatic SNVs and indels in regulatory elements (Nat Genetics, 2022)
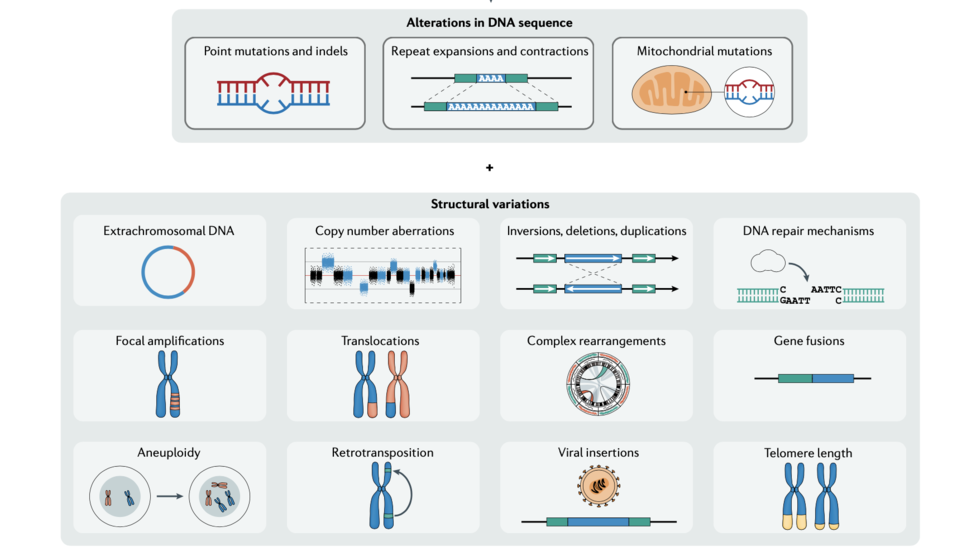
Isidro et al discuss recent advances in cancer genome analysis (Nature Reviews Genetics, 2022)
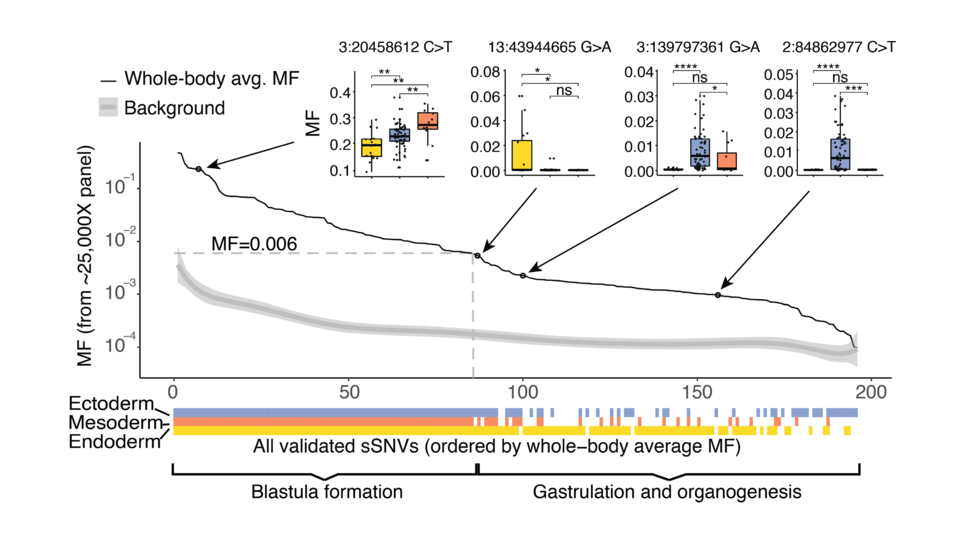
Mosaic variants in multiple organs resolve post-zygotic lineages (Science, 2021)
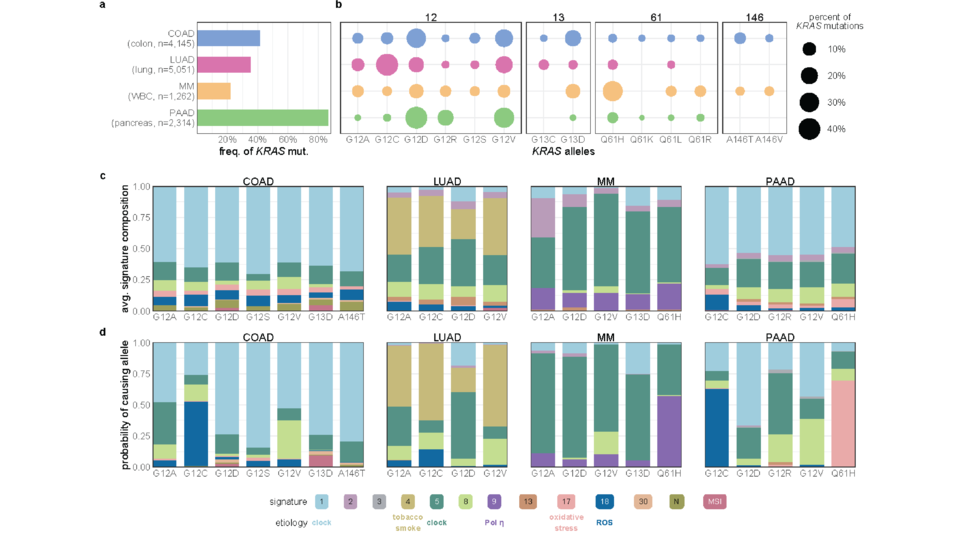
Analysis of KRAS mutations at the allele level using >13K samples (Nat Comm, 2021)
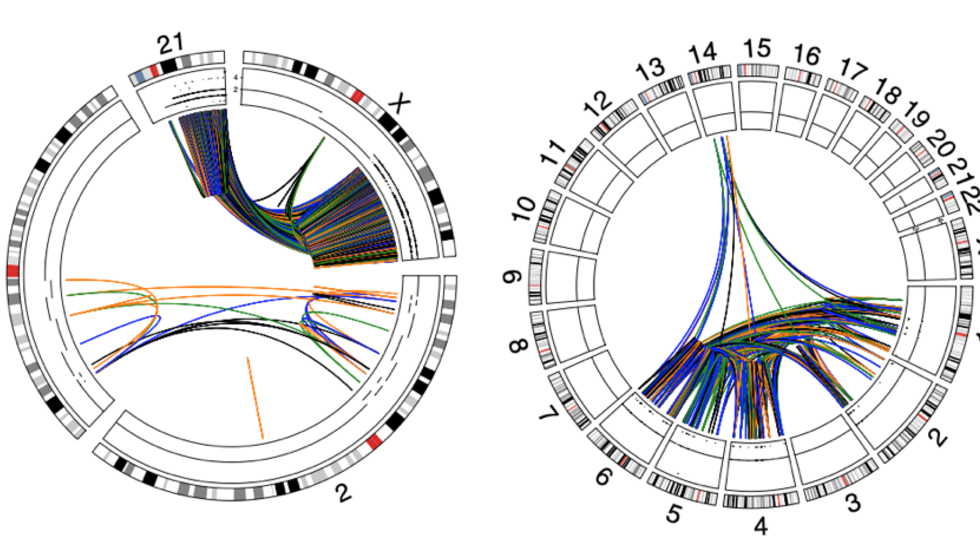
Analysis of >2500 cancer genomes shows that chromothripsis events are prevalent across multiple tumor types (Nat Genetics, 2020)
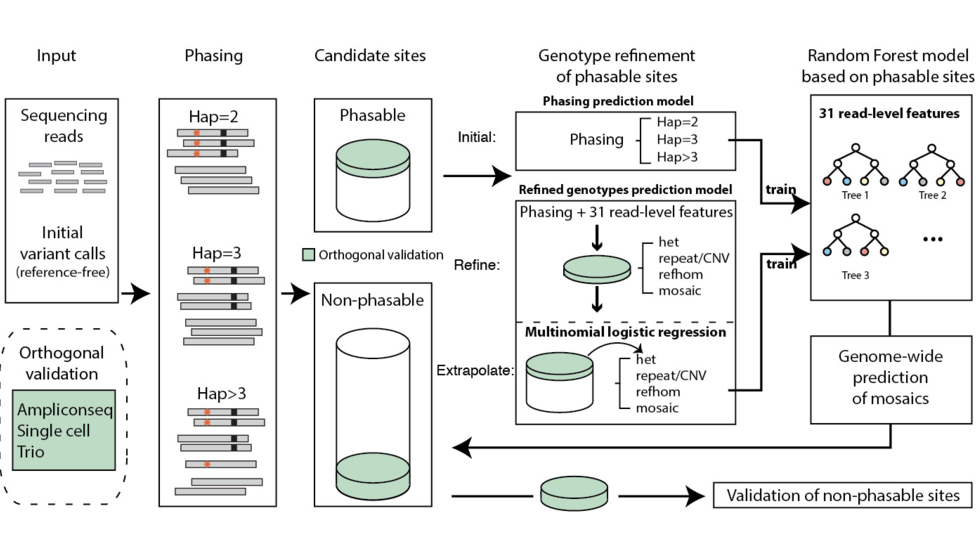
Yanmei et al develop MosaicForecast, a machine learning-based variant caller for mosaic mutations (Nat Biotech, 2020)
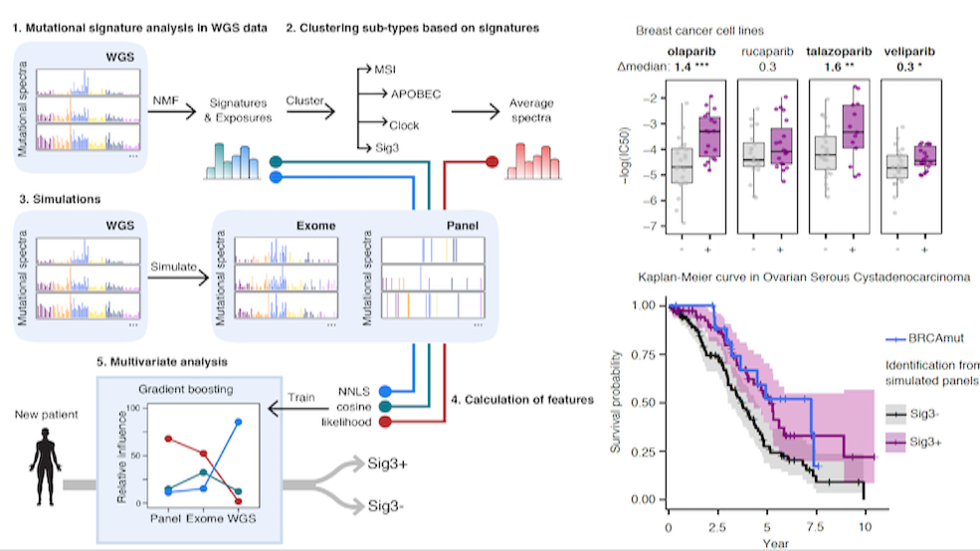
Doga et al develop SigMA to identify patients who may benefit from PARP inhibitor treatment (Nat Genetics, 2019)
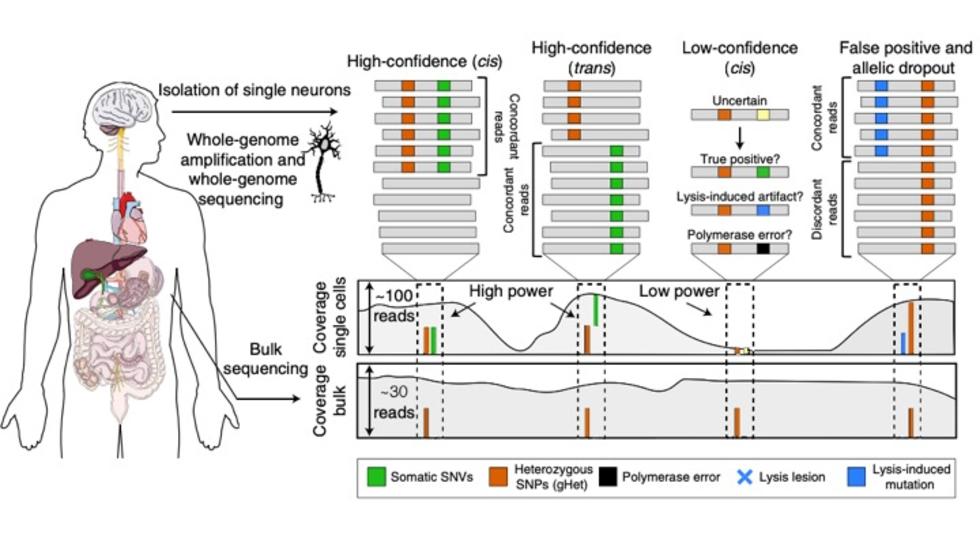
Craig develops a method for detection of somatic variants in single cells (Nat Genetics, 2019)
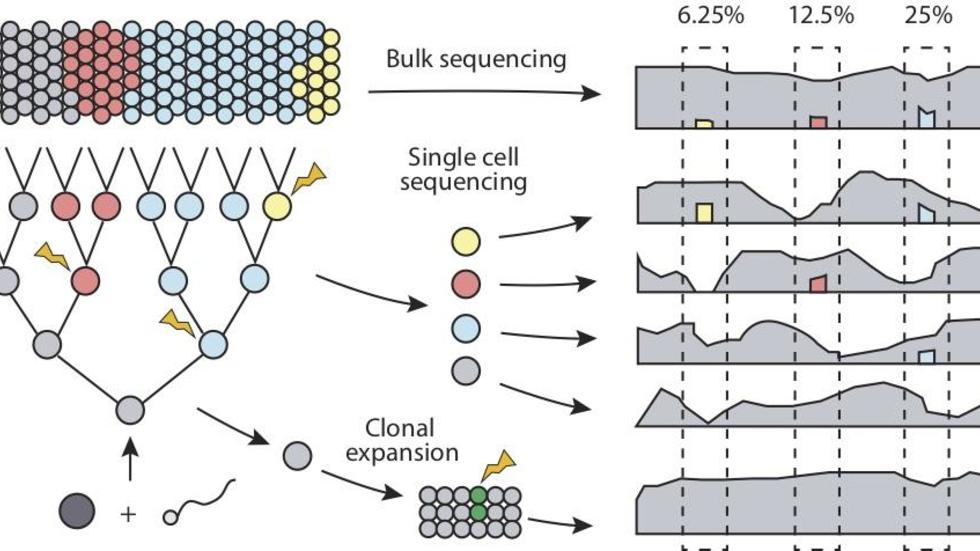
Yanmei, Heather, Joe, and Peter discuss issues in detection of somatic variants (Trends in Genetics, 2018)
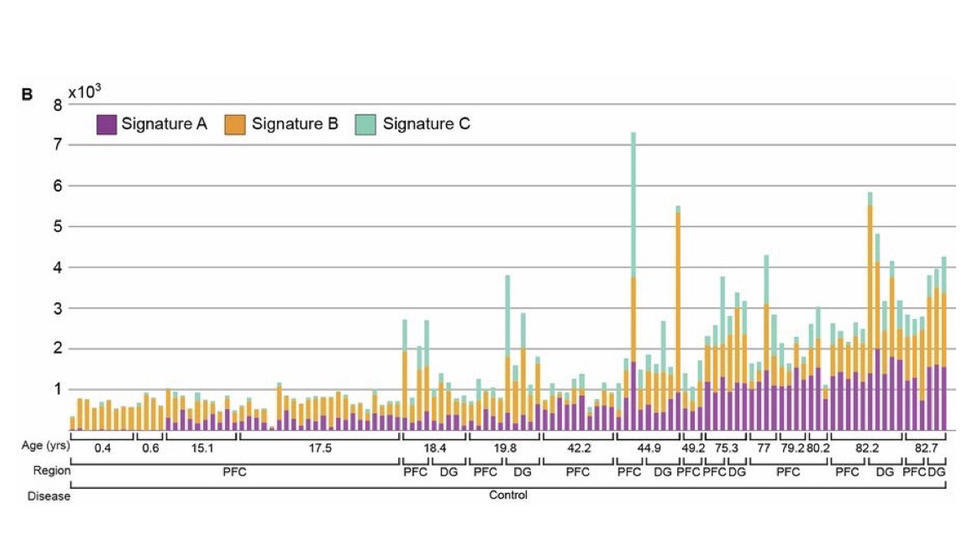
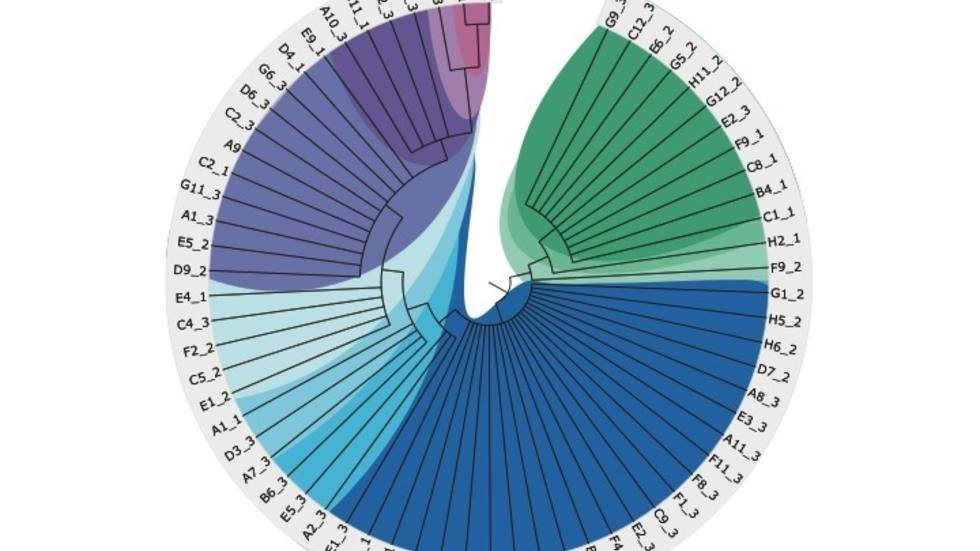
Analysis of the largest single cell WGS dataset to date (Science, 2018)
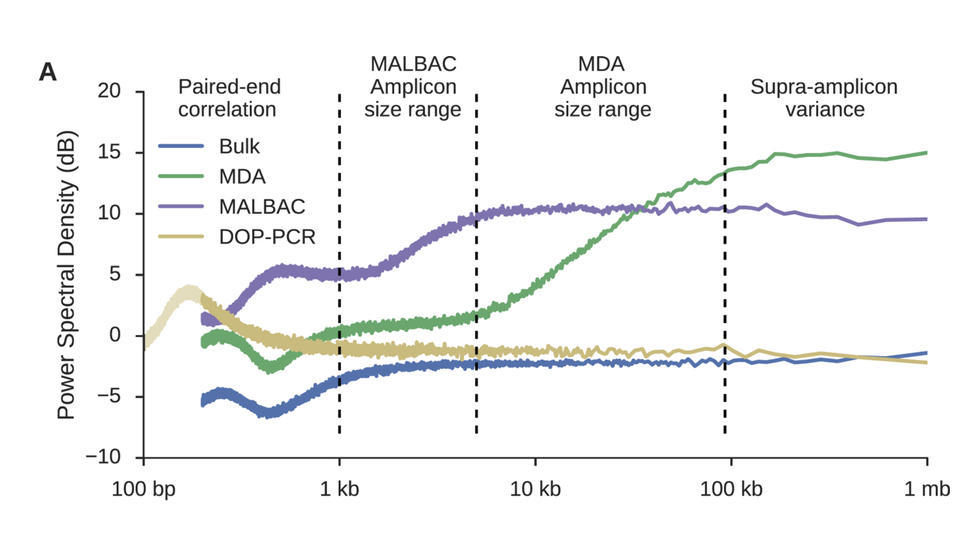
Sherman et al. (Nucleic Acids Research, 2017) develop a QC method based on power density estimation
Cortes-Ciriano et al,Nat Comm, June, 2017
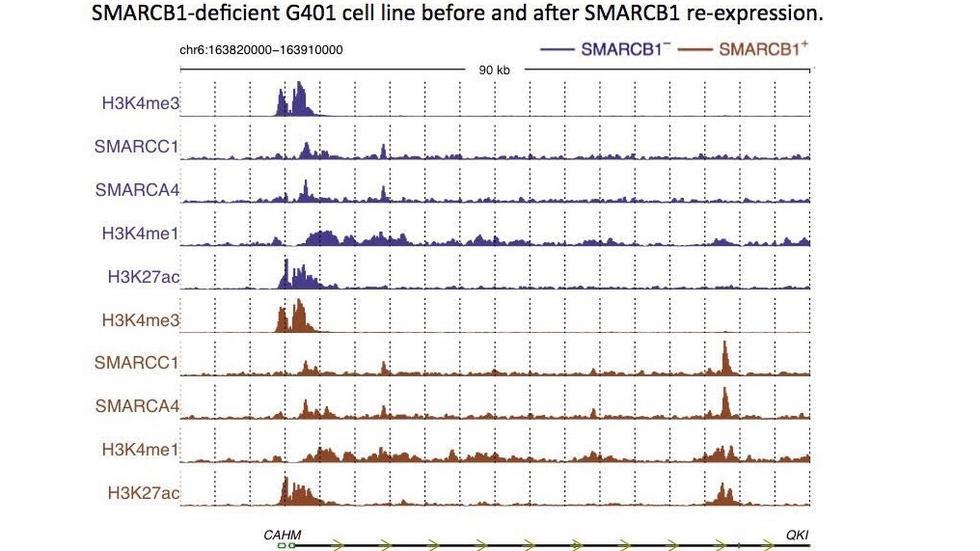
Three papers from a collaboration with the Roberts lab inNature GeneticsandNature Communications
Yang et al (AJHG, 2016) analyze nearly 5000 cancer exomes and identify thousands of rearrangements.
Lineage tracing of neurons using mutational analysis of single cell whole-genome sequencing data (Science, 2015; cover)
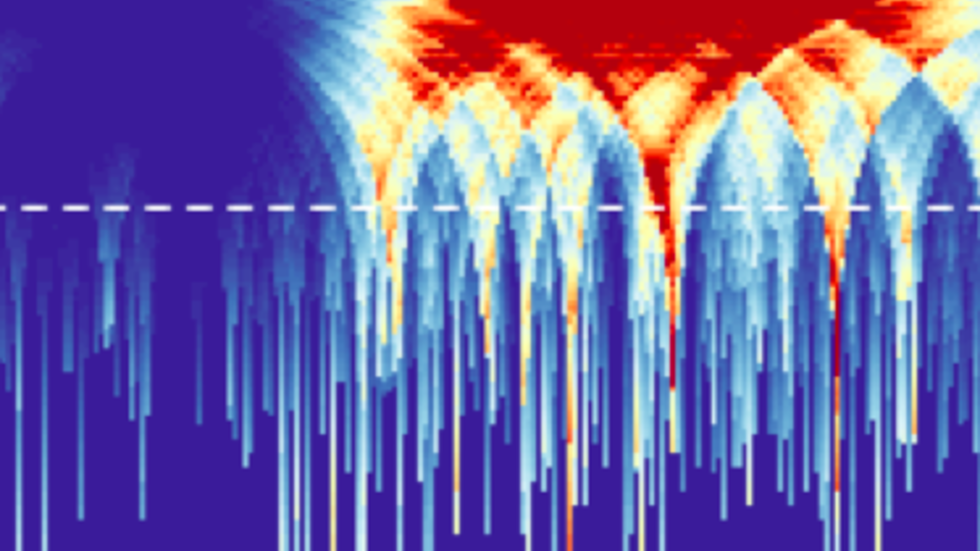
The Park lab, along with the Kharchenko and Gehlenborg labs at DBMI, are leading the Data Coordination and Integration Center
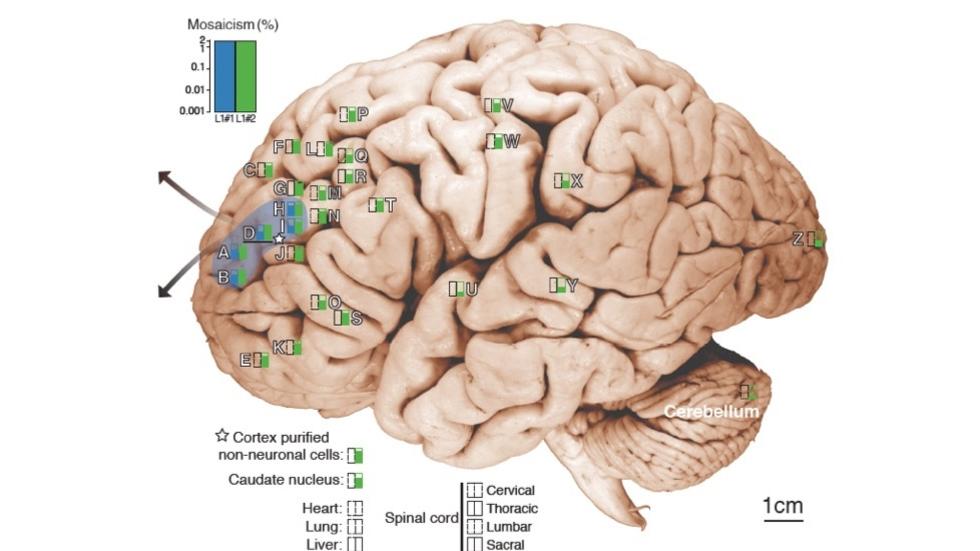
Gilad Evrony, Alice Lee, et al. inNeuron, 2015 (Collaboration with the Chris Walsh lab)
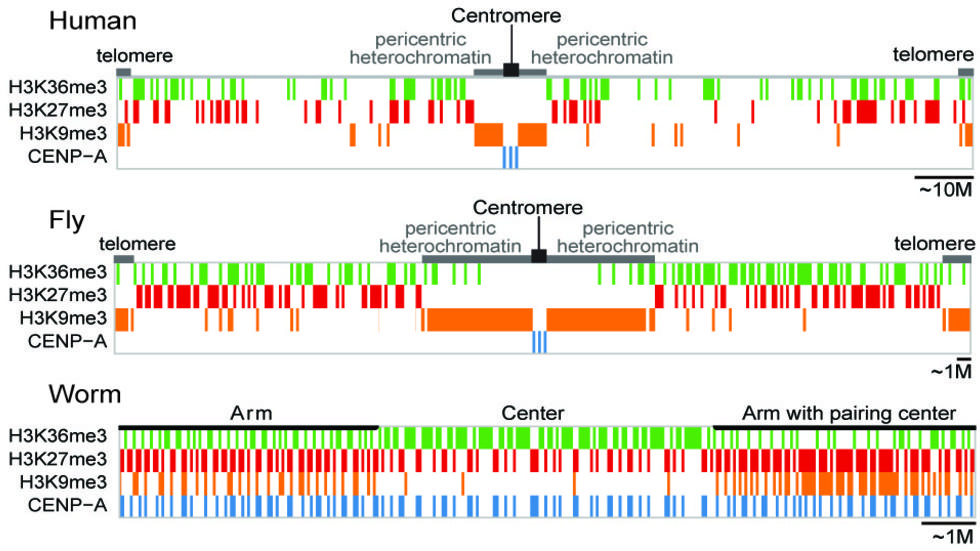
A cross-species analysis from the ENCODE and modENCODE consortia, led by Park lab members,Nature, 2014
We are a bioinformatics research group in the Department of Biomedical Informatics at Harvard Medical School. We are part of the Ludwig Center at Harvard, Division of Genetics at Brigham and Women’s Hospital and Harvard-MIT Division of Health, Science, & Technology.
Our main aim is to understand the genetic and epigenetic mechanisms that underlie disease processes through computational and statistical analysis of genomic data. We are particularly interested in mutational processes in normal and cancer cells and their impact in gene regulation.
We have the privilege of collaborating with a number of superb experimental laboratories at Harvard Medical School and its affiliated hospitals, as well as around the world. Our recent and current collaborators include Connie Cepko, George Daley, Steve Elledge, Konrad Hochedlinger, Mark Johnson, Mitzi Kuroda, Jordan Kreidberg, Bob Kingston, William Pu, Charlie Roberts, Chris Walsh, and Fred Winston.
Latest News
Oct 2025: Lab Hiking Trip 2025
The annual Park lab hiking trip took place on a sunny October Saturday this year. Great views and even greater company!
Aug 2025: Summer Social- Dine Out Boston
Park Lab gathered to try Dine Out Boston (restaurant week) together for our summer social!
Jul 2025: Farewell to Jiny and Niklas!
Park lab said goodbye and good luck to Jiny as she starts her own lab at Hanyang University, and to Niklas as he begins his PhD at EBI. Both will be missed around the lab!
Jun 2025: SMaHT Annual Meeting 2025
As the Data Analysis Center (DAC) of the Somatic Mosaicism Across Human Tissues (SMaHT) Project, Park Lab attended the 2025 Annual Meeting in Bethesda, Maryland.
May 2025: Farewell Clara K!
Park Lab gathered to say goodbye to Clara K on her last day with the team as an Associate Computational Biologist. We wish good luck to Clara Kim on her next adventure in medical school!