Abstract
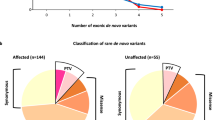
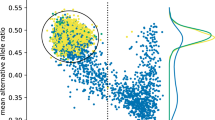
Although germline de novo copy number variants (CNVs) are known causes of autism spectrum disorder (ASD), the contribution of mosaic (early-developmental) copy number variants (mCNVs) has not been explored. In this study, we assessed the contribution of mCNVs to ASD by ascertaining mCNVs in genotype array intensity data from 12,077 probands with ASD and 5,500 unaffected siblings. We detected 46 mCNVs in probands and 19 mCNVs in siblings, affecting 2.8–73.8% of cells. Probands carried a significant burden of large (>4-Mb) mCNVs, which were detected in 25 probands but only one sibling (odds ratio = 11.4, 95% confidence interval = 1.5–84.2, P = 7.4 × 10−4). Event size positively correlated with severity of ASD symptoms (P = 0.016). Surprisingly, we did not observe mosaic analogues of the short de novo CNVs recurrently observed in ASD (eg, 16p11.2). We further experimentally validated two mCNVs in postmortem brain tissue from 59 additional probands. These results indicate that mCNVs contribute a previously unexplained component of ASD risk.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data on individuals with ASD and their families were collected by the Simons Foundation as part of the Simons Simplex Collection and the Simons Powering Autism Research for Knowledge cohort. Mosaic event calls are available in the Supplementary Data. Genotype array data and phenotype information for the SSC and SPARK cohorts are available from SFARI Base (https://base.sfari.org) for approved researchers. Access to the UK Biobank Resource is available via application (http://www.ukbiobank.ac.uk/). Data from the DECIPHER database are available from https://decipher.sanger.ac.uk/. WGS data of postmortem brain tissue are available from the National Institute of Mental Health Data Archive under accession number 1503337. Source data are provided for gels shown in Supplementary Figs. 16c and 17a.
Code availability
MoChA and custom BCFtools plugins are available on Github via URLs listed below. Custom analysis scripts are available from the authors upon reasonable request.
URLs:
MOsaic CHromosomal Alterations (MoChA) caller: https://github.com/freeseek/mocha
BCFtools: https://samtools.github.io/bcftools/bcftools.html
Custom BCFtools plugins: https://github.com/freeseek/gtc2vcf
Eagle2 software: https://data.broadinstitute.org/alkesgroup/Eagle/
PLINK: https://www.cog-genomics.org/plink/1.9/
pyGenomeTracks: https://github.com/deeptools/pyGenomeTracks
1000 Genomes dataset: http://www.1000genomes.org/
Haplotype Reference Consortium: http://www.haplotype-reference-consortium.org/.
UK Biobank: http://www.ukbiobank.ac.uk/
SFARI Gene database: https://gene.sfari.org/
SFARI Base: https://base.sfari.org
References
Gaugler, T. et al. Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885 (2014).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Turner, T. N. et al. Genomic patterns of de novo mutation in simplex autism. Cell 171, 710–722 (2017).
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Sanders, S. J. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885 (2011).
Sanders, S. J. et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015).
Yuen, R. K. C. et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611 (2017).
Iakoucheva, L. M., Muotri, A. R. & Sebat, J. Getting to the cores of autism. Cell 178, 1287–1298 (2019).
McConnell, M. J. et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: the Brain Somatic Mosaicism Network. Science 356, eaal1641 (2017).
Ju, Y. S. et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 543, 714–718 (2017).
Freed, D. & Pevsner, J. The contribution of mosaic variants to autism spectrum disorder. PLoS Genet. 12, e1006245 (2016).
Lim, E. T. et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 20, 1217–1224 (2017).
Krupp, D. R. et al. Exonic mosaic mutations contribute risk for autism spectrum disorder. Am. J. Hum. Genet. 101, 369–390 (2017).
Jamuar, S. S. et al. Somatic mutations in cerebral cortical malformations. N. Engl. J. Med. 371, 733–743 (2014).
Baek, S. T., Gibbs, E. M., Gleeson, J. G. & Mathern, G. W. Hemimegalencephaly, a paradigm for somatic postzygotic neurodevelopmental disorders. Curr. Opin. Neurol. 26, 122 (2013).
Poduri, A., Evrony, G. D., Cai, X. & Walsh, C. A. Somatic mutation, genomic variation, and neurological disease. Science 341, 1237758 (2013).
King, D. A. et al. Mosaic structural variation in children with developmental disorders. Hum. Mol. Genet 24, 2733–2745 (2015).
Fischbach, G. D. & Lord, C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195 (2010).
Feliciano, P. et al. SPARK: a US cohort of 50,000 families to accelerate autism research. Neuron 97, 488–493 (2018).
Loh, P.-R. et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Loh, P.-R. et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 559, 350–355 (2018).
Loh, P.-R., Genovese, G. & McCarroll, S. A. Monogenic and polygenic inheritance become instruments for clonal selection. Nature 584, 136–141 (2020).
Vattathil, S. & Scheet, P. Extensive hidden genomic mosaicism revealed in normal tissue. Am. J. Hum. Genet. 98, 571–578 (2016).
Jacobs, K. B. et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet. 44, 651–658 (2012).
Laurie, C. C. et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 44, 642–650 (2012).
Zaykin, D. V. Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J. Evol. Biol. 24, 1836–1841 (2011).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431 (2019).
Pinto, D. et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 94, 677–694 (2014).
O’Roak, B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
Owen, D. et al. Effects of pathogenic CNVs on physical traits in participants of the UK Biobank. BMC Genomics 19, 867 (2018).
Crawford, K. et al. Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J. Med. Genet. 56, 131–138 (2019).
Bracher-Smith, M. et al. Effects of pathogenic CNVs on biochemical markers: a study on the UK Biobank. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/723270v2 (2019).
McConnell, M. J. et al. Mosaic copy number variation in human neurons. Science 342, 632–637 (2013).
Cai, X. et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 8, 1280–1289 (2014).
Bae, T. et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science 359, 550–555 (2018).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Firth, H. V. et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009).
Riley, K. N. et al. Recurrent deletions and duplications of chromosome 2q11.2 and 2q13 are associated with variable outcomes. Am. J. Med. Genet. A 167A, 2664–2673 (2015).
Forsberg, L. A. Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum. Genet. 136, 657–663 (2017).
Graham, E. J. et al. Somatic mosaicism of sex chromosomes in the blood and brain. Brain Res. 1721, 146345 (2019).
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015).
Nowakowska, B. Clinical interpretation of copy number variants in the human genome. J. Appl Genet. 58, 449–457 (2017).
van den Berg, M. M. J., van Maarle, M. C., van Wely, M. & Goddijn, M. Genetics of early miscarriage. Biochim. Biophys. Acta 1822, 1951–1959 (2012).
Nazeen, S., Palmer, N. P., Berger, B. & Kohane, I. S. Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities. Genome Biol. 17, 228 (2016).
Feenstra, I. et al. Genotype–phenotype mapping of chromosome 18q deletions by high-resolution array CGH: an update of the phenotypic map. Am. J. Med. Genet. Part A 143A, 1858–1867 (2007).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Akbarian, S. et al. The PsychENCODE project. Nat. Neurosci. 18, 1707–1712 (2015).
Wang, M. et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5, 180185 (2018).
Sherman, M. A. et al. PaSD-qc: quality control for single cell whole-genome sequencing data using power spectral density estimation. Nucleic Acids Res. 46, e20 (2018).
Feliciano, P. et al. Exome sequencing of 457 autism families recruited online provides evidence for novel ASD genes. NPJ Genom. Med. 4, 19 (2019).
Diskin, S. J. et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 36, e126 (2008).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007).
Newcombe, R. G. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat. Med. 17, 873–890 (1998).
Loh, P.-R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906 (2018).
Jacquemont, S. et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 478, 97–102 (2011).
Dong, S. et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 9, 16–23 (2014).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Genovese, G., Handsaker, R. E., Li, H., Kenny, E. E. & McCarroll, S. A. Mapping the human reference genome’s missing sequence by three-way admixture in Latino genomes. Am. J. Hum. Genet. 93, 411–421 (2013).
Evrony, G. D. et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell 151, 483–496 (2012).
Baslan, T. et al. Genome-wide copy number analysis of single cells. Nat. Protoc. 7, 1024–1041 (2012).
Ramírez, F. et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 1–15 (2018).
Acknowledgements
We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren and E. Wijsman). We are grateful to all of the families in SPARK, the SPARK clinical sites and SPARK staff. We appreciate obtaining access to genotype and phenotype data on SFARI Base. Approved researchers can obtain the SSC and SPARK population dataset described in this study by applying at https://base.sfari.org/. We would like to thank the HMS Research Computing Consultant Group for their consulting services, which facilitated the computational analyses detailed in this article. This research was conducted using the UK Biobank Resource under application no. 19808. M.A.S. is supported by a grant from the NIMH under award no. F31MH124393. R.E.R. is supported by the Stuart H.Q. and Victoria Quan Fellowship in Neurobiology and by the Harvard/MIT MD–PhD program (T32GM007753) from the NIGMS. G.G. was supported by NIH grant R01HG006855, NIH grant R01MH104964 and the Stanley Center for Psychiatric Research. C.M.D is supported by the NIMH Translational Post-doctoral Training Program in Neurodevelopment (T32MH112510). A.R.B. was supported by training grant T32HG229516 from the NHGRI. R.E.M. is supported by NSF grant DMS-1939015 and NIH grant K25HL150334. B.B. is supported by grant R01GM108348 from the NIGMS. P.J.P is supported by NIMH grant U01MH106883 and the Harvard Ludwig Center. C.A.W. is supported by the Allen Discovery Center program through the Paul G. Allen Frontiers Group and grants from the NINDS (R01NS032457) and the NIMH (U01MH106883) C.A.W. is an Investigator of the Howard Hughes Medical Institute. P.-R.L. is supported by NIH grant DP2 ES030554, a Burroughs Wellcome Fund Career Award at the Scientific Interfaces, the Next Generation Fund at the Broad Institute of MIT and Harvard, the Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty award and a Sloan Research Fellowship. WGS data were generated as part of the Brain Somatic Mosaicism Network Consortium. A full list of supporting grants and consortium members are provided in the Supplementary Information. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
M.A.S., P.J.P., C.A.W. and P.-R.L. conceived and designed the study. M.A.S., G.G. and P.-R.L. designed and implemented the statistical methods. M.A.S. performed computational analyses. C.D. curated phenotype data. R.E.R. performed WGS and experimental validation in postmortem brain tissue. A.R.B., R.E.M. and B.B. provided comments and guidance throughout. All authors wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Ethics statement
The first part of this study used existing and publicly available genomic datasets of families with ASD from the Simons Simplex Collection (SSC) and Simons Powering Autism Research for Knowledge (SPARK). Collection of SSC samples was approved and monitored by the institutional review board of Columbia University Medical Center. SPARK samples were collected under a centralized review board protocol (Western IRB Protocol no. 20151664). The second part of the study generated and analyzed genomic data on de-identified postmortem human specimens obtained from brain tissue banks, including the AutismBrainNet, the Lieber Institute for Brain Development, the Oxford Brain Bank and the University of Maryland Brain and Tissue Bank through the National Institutes of Health Neurobiobank. This study did not engage human subjects or collect their identifiable data; rather, the individual tissue banks have their own approval and consent process. Our study was approved by the institutional review board of Boston Children’s Hospital.
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Carrie Bearden, Stephan Sanders and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–23 and Supplementary Notes 1–14.
Supplementary Tables
Supplementary Tables 1–15.
Supplementary Data
Original, unmodified gels corresponding to those shown in Supplementary Fig. 17a.
Rights and permissions
About this article
Cite this article
Sherman, M.A., Rodin, R.E., Genovese, G. et al. Large mosaic copy number variations confer autism risk. Nat Neurosci 24, 197–203 (2021). https://doi.org/10.1038/s41593-020-00766-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00766-5
This article is cited by
-
Increased cerebral lactate levels in adults with autism spectrum disorders compared to non-autistic controls: a magnetic resonance spectroscopy study
Molecular Autism (2023)
-
Facilitating genome structural variation analysis
Nature Methods (2023)
-
Somatic genetic variation in healthy tissue and non-cancer diseases
European Journal of Human Genetics (2023)
-
Deep exome sequencing identifies enrichment of deleterious mosaic variants in neurodevelopmental disorder genes and mitochondrial tRNA regions in bipolar disorder
Molecular Psychiatry (2023)
-
Genomic Mosaicism of the Brain: Origin, Impact, and Utility
Neuroscience Bulletin (2023)